Research
Overview of Research Expertise
I have a broad research portfolio that spans bacterial pathogenesis, vaccine development (TB and Influenza), host-pathogen interactions, 3-D organotypic cell culture models, multispecies microbial communities, and space microbiology. My research interests focus on understanding how microorganisms survive, adapt, and evolve in hostile environments—whether in spaceflight conditions or during infection within human hosts—with the overarching aim of improving human health and developing novel therapeutic strategies.
My interdisciplinary work is critical to addressing challenges in areas such as multidrug-resistant (MDR) infections, spaceflight biofilms, and pathogens requiring T-cell immunity, contributing to advancements in both healthcare and space exploration. Currently, I am focused on three key research areas: 1) understanding the mechanosensing mechanisms during multidrug-resistant (MDR) infections that could lead to systemic infection, 2) exploring how microbial ecological success contributes to persistence and pathogenicity, and 3) investigating how space environments influence microbial behaviors and host-pathogen interactions, with implications for human health in space.
Research Interests
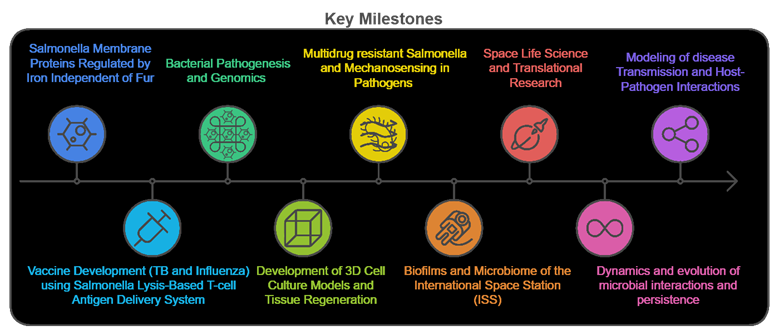
Salmonella Membrane Proteins Regulated by Iron Independent of Fur
From 2003 to 2005, my research focused on Salmonella Typhimurium, investigating membrane proteins and their roles in iron regulation and pathogenesis. Notably, I identified and characterized OmpW, an outer membrane protein expressed under high iron conditions, independent of the Fur regulatory mechanism. My work revealed critical role of OmpW in maintaining bacterial membrane integrity under iron stress and its contribution to Salmonella survival in iron-rich environments. I also explored how these mechanisms influence bacterial adhesion and virulence in host environments, contributing to a deeper understanding of Salmonella pathogenesis.
Key Publications:
- Yoo, A.Y., Yu, J. E., Yang, J, Kim, Y. H., Baek, C. H., Oh, J. I., Kang, H. Y. (2008) Regulation of an outer membrane protein, OmpW, expression and its biological function in Salmonella Typhimurium. Kor J Life Sci 18(11): 1606-1611.
Vaccine Development (TB and Influenza) Using Salmonella Lysis-Based T-cell Antigen Delivery System
From 2008 to 2012, my research focused on vaccine development using a lysis-based antigen delivery system within Live Attenuated Salmonella Vaccines (RASV). I engineered Salmonella strains with regulated delayed attenuation, antigen synthesis, and lysis to release protective antigens, inducing robust immune responses such as CD8+ cytotoxic T-cell responses. This system was applied to vaccines for TB and influenza, showing significant promise in enhancing immune protection.
The vaccines improved efficacy for both TB and influenza, triggering Th1-type immune responses and robust CD8+ T-cell responses, which are critical for viral or TB clearance. My research demonstrated that these lysis-based vaccines offer a safer and more effective alternative for delivering antigens, particularly for challenging pathogens like TB and influenza, contributing to advancements in vaccine technology.
Key Publications:
- Ashraf, S., Kong, W., Wang, S. F., Yang, J. S. and Curtiss, R. (May 23 2011) "Protective cellular responses elicited by vaccination with influenza nucleoprotein delivered by a live recombinant attenuated Salmonella vaccine", Vaccine, 29(23), pp. 3990-4002. doi:10.1016/j.vaccine.2011.03.066 (IF 3.49)
- Juarez-Rodriguez, M. D., Yang, J., Kader, R., Alamuri, P., Curtiss, R. and Clark-Curtiss, J. E. (Feb 2012) "Live Attenuated Salmonella Vaccines Displaying Regulated Delayed Lysis and Delayed Antigen Synthesis To Confer Protection against Mycobacterium tuberculosis", Infection and Immunity, 80(2), pp. 815-831. doi:10.1128/Iai.05526-11 (IF 3.256)
Bacterial Pathogenesis and Genomics
My research in bacterial pathogenesis and genomics focuses on understanding the genetic mechanisms that influence virulence and immune response. This work involves analyzing genes responsible for lipopolysaccharide synthesis and other key virulence factors in Salmonella enterica, as well as conducting comparative genomic analyses to uncover insights into pathogenicity and host interactions.
Key Publications:
- Kong, Q. K., Yang, J. S., Liu, Q., Alamuri, P., Roland, K. L. and Curtiss, R. (Oct 2011) "Effect of Deletion of Genes Involved in Lipopolysaccharide Core and O-Antigen Synthesis on Virulence and Immunogenicity of Salmonella enterica Serovar Typhimurium", Infection and Immunity, 79(10), pp. 4227-4239. doi:10.1128/Iai.05398-11 (IF 3.256)
- Luo, Y. Q., Kong, Q. K., Yang, J., Golden, G., Wanda, S. Y., Jensen, R. V., Ernst, P. B. and Curtiss, R. (Aug 2011) "Complete Genome Sequence of the Universal Killer Salmonella enterica Serovar Typhimurium UK-1 (ATCC 68169)", Journal of Bacteriology, 193(15), pp. 4035-4036. doi:10.1128/Jb.05224-11 (IF 3.825)
- Luo, Y. Q., Kong, Q. K., Yang, J., Mitra, A., Golden, G., Wanda, S. Y., Roland, K. L., Jensen, R. V., Ernst, P. B. and Curtiss, R. (Jul 2012) "Comparative Genome Analysis of the High Pathogenicity Salmonella Typhimurium Strain UK-1", Plos One, 7(7). doi:ARTN e4064510.1371/journal.pone.0040645 (IF 4.41)
- Koroli, S., Buss, K., Blain, J. M., Nakka, G. S., Hong, M., McLean, R. J., Plugge, C. M. and Yang, J†. (Jan 17 2024) "Draft genome sequence of Sphingomonas paucimobilis strain Sph5, isolated from tap water filtration membrane", Microbiol Resour Announc, 13(1), pp. e0034523. doi:10.1128/MRA.00345-23 †Corresponding author
Development of 3D Cell Culture Models and Tissue Regeneration
I developed 3D organotypic cell culture models co-cultured with immune cells to study microbial survival and host-pathogen interactions in physiologically relevant conditions. These models provide critical insights into infection dynamics and immune responses.
Key Publications:
- Barrila, J., Crabbé, A., Yang, J., Franco, K., Nydam, S. D., Forsyth, R. J., Davis, R. R., Gangaraju, S., Ott, C. M., Coyne, C. B., Bissell, M. J. and Nickerson, C. A. (Nov 2018) "Modeling Host-Pathogen Interactions in the Context of the Microenvironment: Three-Dimensional Cell Culture Comes of Age", Infect Immun, 86(11). e00282-18 doi:10.1128/IAI.00282-18 (IF 3.256)
- Barrila, J., Yang, J. (Co-first), Crabbé, A., Sarker, S. F., Liu, Y., Ott, C. M., Nelman-Gonzalez, M. A., Clemett, S. J., Nydam, S. D., Forsyth, R. J., Davis, R. R., Crucian, B. E., Quiriarte, H., Roland, K. L., Brenneman, K., Sams, C., Loscher, C. and Nickerson, C. A. (Feb 28 2017) "Three-dimensional organotypic co-culture model of intestinal epithelial cells and macrophages to study Salmonella enterica colonization patterns", NPJ Microgravity, 3, pp. 10. doi:10.1038/s41526-017-0011-2 (IF 5.1)
MDR invasive Salmonella and Mechanosensing in Pathogens
I demonstrated how fluid shear regulates the virulence and stress responses of pathogens like Salmonella Typhimurium, revealing how spaceflight-relevant low-shear environments or high fluid shear during systemic infections influence microbial behavior and virulence. Particularly, I investigated multidrug-resistant Salmonella ST313, discovering that fluid shear enhances pathogenesis by upregulating key virulence genes and promoting infection in human cells.
Key Publications:
- Yang, J., Barrila, J. (co-first), Roland, K. L., Ott, C. M. and Nickerson, C. A. (Jun 2016) "Physiological fluid shear alters the virulence potential of invasive multidrug-resistant non-typhoidal Salmonella Typhimurium D23580", NPJ Microgravity, 2, pp. 16021. doi:10.1038/npjmgrav.2016.21 (IF 5.1)
- Yang, J., Barrila, J., Roland, K. L., Kilbourne, J., Ott, C. M., Forsyth, R. J. and Nickerson, C. A. (Jun 2015) "Characterization of the Invasive, Multidrug Resistant Non-typhoidal Salmonella Strain D23580 in a Murine Model of Infection", PLoS Negl Trop Dis, 9(6), pp. e0003839. doi:10.1371/journal.pntd.0003839 (IF 3.256)
- Yang J.†, Barrila J, Nauman EA, Nydam SD, Yang S, Jin Park, Gutierrez-Jensen AD, Castro CL, Mark Ott C, Buss K, Steel J, Zakrajsek AD, Schuff MM, Nickerson CA†. (May 2024) "Incremental increases in physiological fluid shear progressively alter pathogenic phenotypes and gene expression in multidrug resistant Salmonella", Gut Microbes, DOI - 10.1080/19490976.2024.2357767 †Corresponding author (IF 12.2)
- Franco Meléndez, K., Crenshaw, K., Barrila, J., Yang, J., Gangaraju, S., Davis, R. R., Forsyth, R. J., Ott, C. M., Kader, R., Curtiss, R., Roland, K. and Nickerson, C. A. (Aug 31 2022) "Role of RpoS in Regulating Stationary Phase Salmonella Typhimurium Pathogenesis-Related Stress Responses under Physiological Low Fluid Shear Force Conditions", mSphere, 7(4), pp. e0021022. doi:10.1128/msphere.00210-22 (IF 4.17)
- Barrila, J., Yang, J., Franco Meléndez, K. P., Yang, S., Buss, K., Davis, T. J., Aronow, B. J., Bean, H. D., Davis, R. R., Forsyth, R. J., Ott, C. M., Gangaraju, S., Kang, B. Y., Hanratty, B., Nydam, S. D., Nauman, E. A., Kong, W., Steel, J. and Nickerson, C. A. (May 31 2022) "Spaceflight Analogue Culture Enhances the Host-Pathogen Interaction Between Salmonella and a 3-D Biomimetic Intestinal Co-Culture Model", Front Cell Infect Microbiol, 12, pp. 705647. doi:10.3389/fcimb.2022.705647 (IF 6.073)
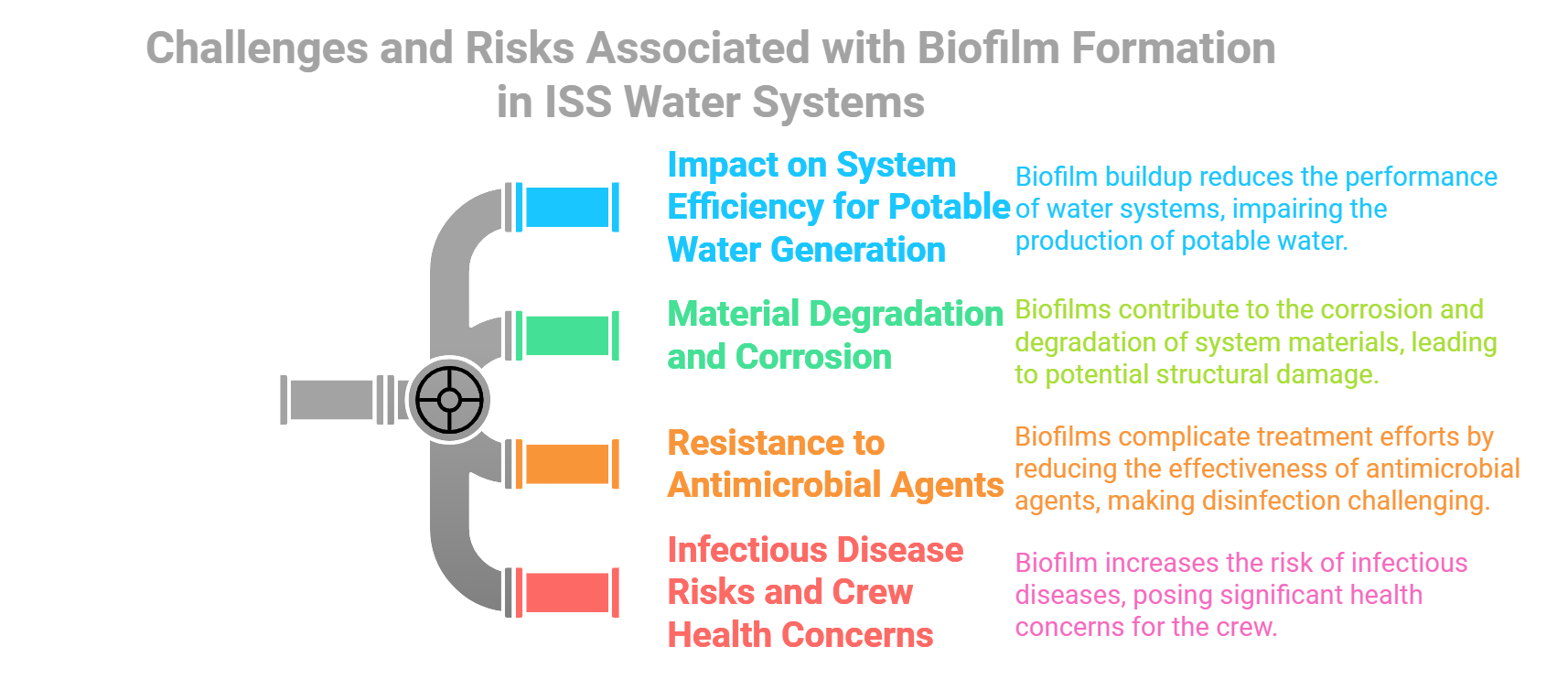
Biofilms and Microbiome of the International Space Station (ISS)
My research on the ISS microbiome focuses on characterizing biofilms and multispecies microbial populations in spaceflight environments, which provides insights into microbial survival, adaptation, and interactions under spaceflight conditions. This work is critical for understanding how to manage microbial communities to ensure crew health and equipment integrity.

Key Publications:
- Yang, J†., Barrila, J., Mark Ott, C., King, O., Bruce, R., McLean, R. J. C. and Nickerson, C. A.† (Sep 06 2021) "Longitudinal characterization of multispecies microbial populations recovered from spaceflight potable water", NPJ Biofilms Microbiomes, 7(1), pp. 70. doi:10.1038/s41522-021-00240-5 †Corresponding author (IF 8.462)
- Yang, J., Thornhill, S. G., Barrila, J., Nickerson, C. A., Ott, C. M. and McLean, R. J. C. (2018) "Microbiology of the Built Environment in Spacecraft Used for Human Flight", Microbiology of Atypical Environments, 45, pp. 3-26. doi:10.1016/bs.mim.2018.07.002
- Vélez Justiniano, Y. A., Goeres, D. M., Sandvik, E. L., Kjellerup, B. V., Sysoeva, T. A., Harris, J. S., Warnat, S., McGlennen, M., Foreman, C. M., Yang, J., Li, W., Cassilly, C. D., Lott, K. and HerrNeckar, L. E. (Dec 2023) "Mitigation and use of biofilms in space for the benefit of human space exploration", Biofilm, 5, pp. 100102. doi:10.1016/j.bioflm.2022.100102 (IF 5.27)
Space Microbiology
Since 2015, my research has focused on spaceflight microbiology and microbial long-term adaptation and host-pathogen interactions in space. My research has expanded to characterizing multispecies microbial communities in the ISS potable water system, supported by the Alfred P. Sloan Foundation's MoBE program. I investigated microbial interactions and adaptation, identifying microevolution in space and key interactions influencing system integrity and astronaut health. My ongoing studies explore microbial interactions and adaptation during long-duration spaceflight, providing further insight into microbial dynamics and risks to crew health.
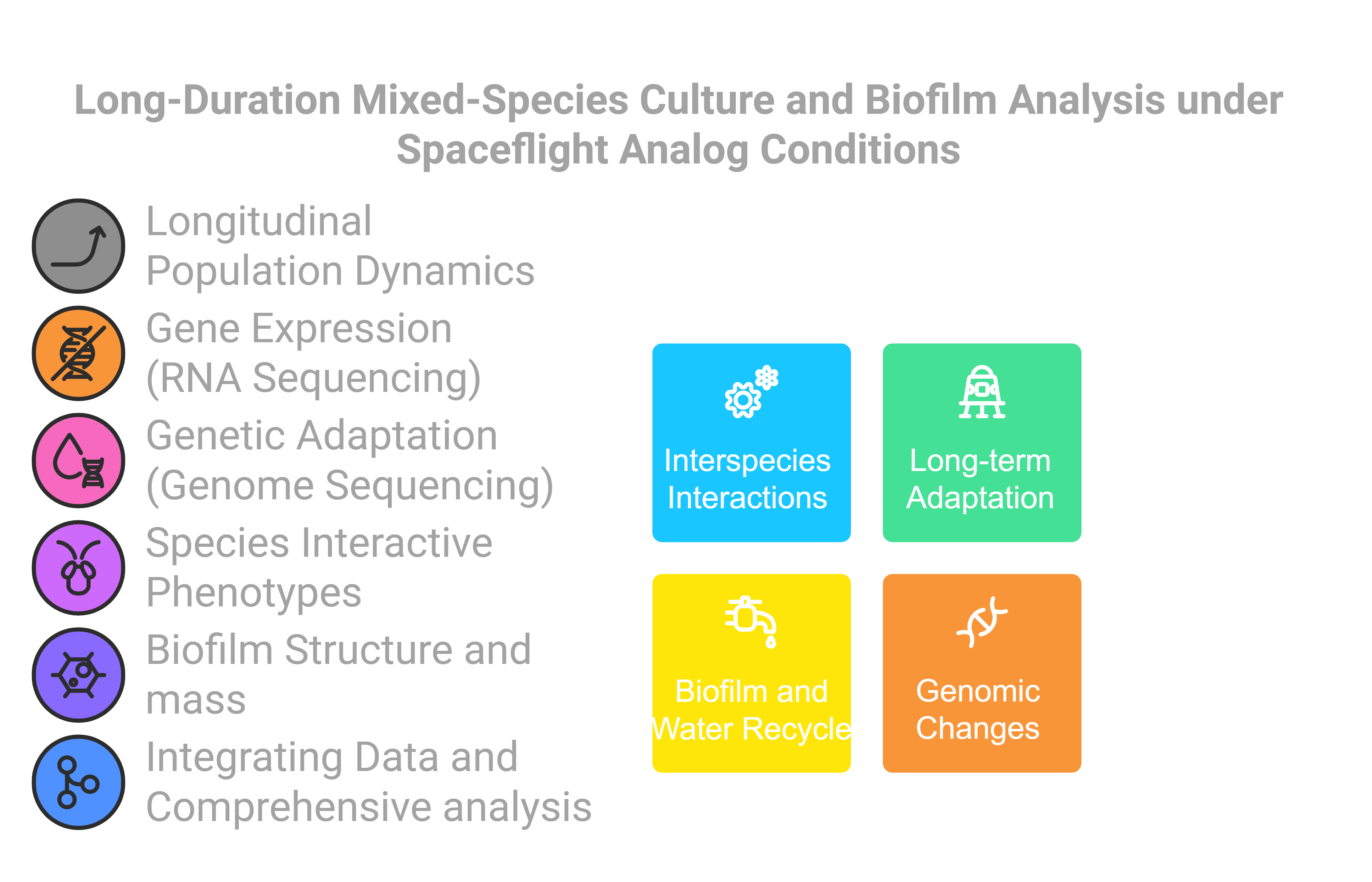
Micro-17 is an ongoing project investigating the population dynamics, species interactions, biofilm formation, gene expression, and genetic adaptations of microbial communities over 100 days under spaceflight-analog conditions. Initially funded as a spaceflight experiment to explore the evolution of species interactions, the project shifted to a ground-based study due to hardware development challenges, while retaining its original scope. This research examines adaptive responses and genetic shifts within heterogeneous populations, comparing microbial interactions and resilience in simulated spaceflight versus standard gravity (1xg) environments, with implications for microbial resilience in space.
The Micro-17 project addresses critical gaps in understanding microbial interactions, resilience, and evolution, with broad implications for both space exploration and terrestrial applications. By examining how prolonged exposure to a spaceflight-analog condition influences microbial community dynamics, resilience, and genetic adaptation over time, this research offers valuable insights into microbial ecology, applied microbiology, and evolutionary biology in extreme conditions. Findings from this study not only inform strategies to mitigate microbial risks in long-term space missions but also enhance our understanding of microbial interactions, resilience, and evolution potential in confined or challenging settings on Earth, such as healthcare facilities and industrial systems.
Key Publications:
- Santomartino, R., Averesch, N. J. H., Bhuiyan, M., Cockell, C. S., Colangelo, J., Gumulya, Y., Lehner, B., Lopez-Ayala, I., McMahon, S., Mohanty, A., Santa Maria, S. R., Urbaniak, C., Volger, R., Yang, J. and Zea, L. (Mar 21 2023) "Toward sustainable space exploration: a roadmap for harnessing the power of microorganisms", Nat Commun, 14(1), pp. 1391. doi:10.1038/s41467-023-37070-2 *Authors alphabetical ordered, equally contributed (IF 17.7)
Dynamics and evolution of microbial interactions and persistence
Modeling of disease Transmission and Host-Pathogen Interactions